
Which annotation features could be used to rank the mutation list for a clinician?.How could we use variation archives to further filter the list of exonic variants?.What would be a useful additional filter in our case given that the index patient has consanguineous parents?.īcftools query -f "%CHROM\t%POS\t%ID\t%REF\t%ALT\t\n" Make sure you use the hg19/GRCh37 version available here. Then copy and paste into the VEP application. We first dump all SNPs in a VEP compliant format which you can In this practical we will use VEP because it can be run directly online. In the recent years a number of convenient pipelines have been developed that ease the annotation of variants with some of the above information. you can categorize known mutations into benign and pathogenic using ClinVar.you can prioritize mutations in genes that interact with known candidate genes of the disease.you can check the expression of genes in your studied tissue using GTEx.you can annotate variants with allele frequency information from variation archives such as 1000 Genomes, ExAC or gnomAD.there is a long list of mutation damaging prediction tools such as PolyPhen, MutationTaster or Sift.you can use transcript annotations from Ensembl, UCSC or RefSeq.
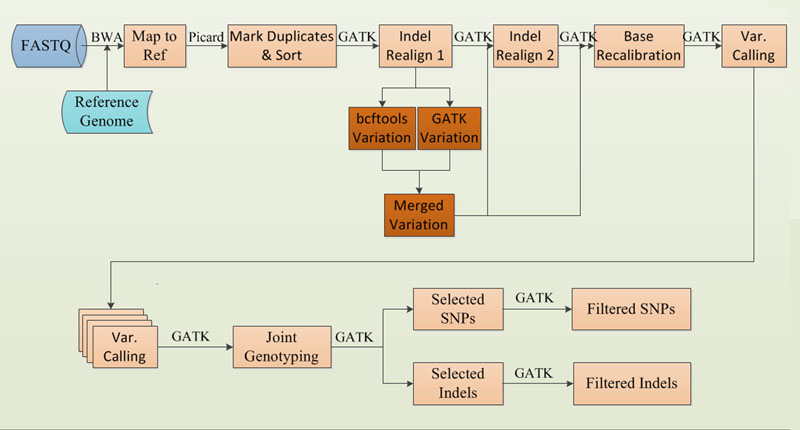
Variant annotation and classification is a challenging process.
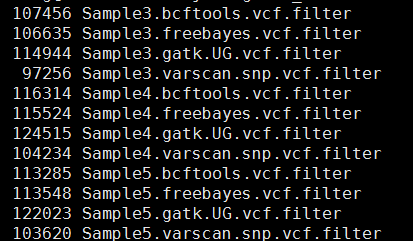
What is the genotype and the allelic depth for both variants that FreeBayes emits?.Are the first two exonic variants homozygous or heterozygous?.Samtools tview -d t -p chr7:299825 rd.rmdup.bam chr7.fa


 0 kommentar(er)
0 kommentar(er)
